Detector response to a high-energy physics process is often estimated by Monte Carlo simulation. For purposes of data analysis, the results of this simulation are typically stored in large multi-dimensional histograms, which can quickly become unwieldy in terms of size or numerically problematic due to unfilled bins or interpolation artifacts. Photospline is a library that uses the penalized spline technique to efficiently compute, store, and evaluate B-spline representations of such tables.
A basic fitting script starts with a few imports:
import numpy
from icecube.photospline import spglam as glam
from icecube.photospline import splinefitstable
from icecube.photospline.glam.glam import grideval
from icecube.photospline.glam.bspline import bspline
Some of these are strictly required.
We will also need some extra functions in order to visualize the result.
For this example, we will make up some random, one-dimensional data drawn from a cosine overlaid on a polynomial:
numpts = 500
x1 = numpy.sort(numpy.random.uniform(-4,25,size=numpts))
z = numpy.random.poisson(numpy.cos(x1)**2 + (x1-3.0)**2 + 10)
We can improve the quality of the fit by giving less weight to data points that are likely to be distorted by statistical fluctuations. Here we’ll use a minimum weight of 1, and weight the high-occupancy points up:
w = 1. + z
Now we need to choose a set of B-spline basis functions. We will use a grid of order-2 B-splines that extend beyond the the data points, relying on the regularization term for extrapolation:
order = 2
knots = [numpy.linspace(-8,35,30)]
smooth = 3.14159e3
This generalizes easily to multi-dimensional data; we would simply add an extra entry to knots for each extra dimension.
To actually run the fit, we call icecube.photospline.spglam.fit()
>>> result = glam.fit(z,w,[x1],knots,order,smooth)
Calculating penalty matrix...
Calculating spline basis...
Reticulating splines...
Convolving bases...
Convolving dimension 0
Flattening residuals matrix...
Transforming fit array...
Computing least square solution...
Analyze[27]: 0.000049 s
Factorize[27]: 0.000027 s
Solve[27]: 0.000022 s
Done: cleaning up
Now we can save the fit result for later use with ndsplineeval() or icecube.photospline.glam.glam.grideval()
splinefitstable.write(result, 'splinefit-1d.fits')
To see the result, we can plot it with matplotlib:
import pylab
# Plot individual basis splines
xfine = numpy.linspace(knots[0][0], knots[0][-1], 10001)
splines = [numpy.array([bspline(knots[0], x, n, order) for x in xfine]) for n in range(0,len(knots[0])-2-1)]
for c, n in colorize(range(len(splines))):
pylab.plot(xfine, result.coefficients[n]*splines[n], color=c)
# Plot the spline surface (sum of all the basis functions)
pylab.plot(xfine, glam.grideval(result, [xfine]), label='Spline fit', color='k')
pylab.legend(loc='upper left')
pylab.show()
The result is shown below:
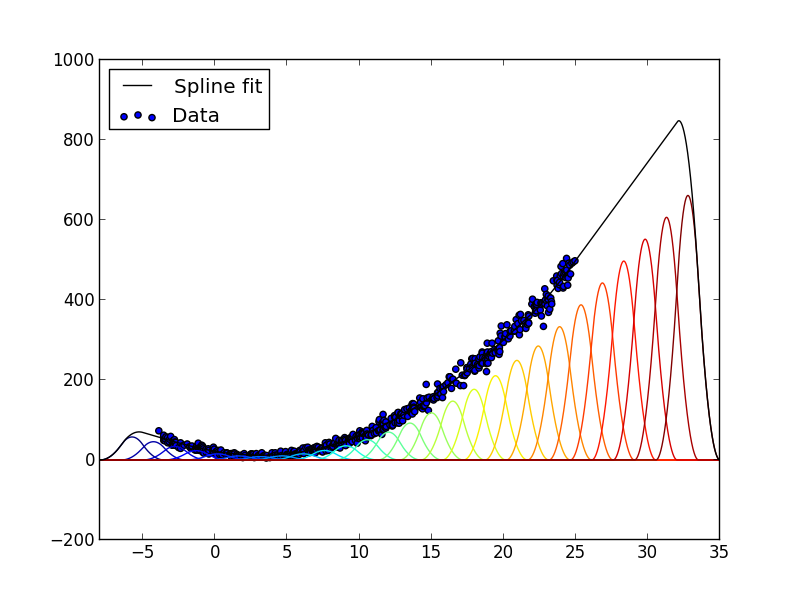
Example 1-dimensional spline fit. The spline surface (black line) is the sum of basis functions (colored lines) weighted with coefficients determined by a linear least-squares fit to the data (blue dots). In the region to the right where there are no data, the order-2 penalty term produces a straight line.
The interface to the fitting library is entirely in Python, using Numpy arrays to as containers for the data, spline coefficients, knot vectors, etc. The spline coefficients determined in the fit are stored in FITS files that can be loaded and evaluated using the bundled C library.
Note
The reference Python fitting implementation icecube.photospline.glam.glam.fit() uses dense matrix operations from numpy, and can become extremely memory-hungry in large numbers of dimensions.
The C implementation icecube.photospline.spglam.fit() implements the same functions as the Python version, but uses sparse matrix operations from SuiteSparse. This is both orders of magnitude faster and uses 100x less memory for typical problems. You should use spglam for large-scale fitting and fall back to glam if necessary for debugging.
Fit gridded data to a B-spline surface.
| Parameters: |
|
|---|---|
| Returns: | SplineTable - the fit result |
A drop-in replacement for icecube.photospline.glam.glam.fit().
Write a SplineTable to disk as a FITS file.
| Parameters: |
|
|---|
Warning
pyfits will fail to write the file if it already exists.
Read a SplineTable from a FITS file on disk
| Parameters: | path – the filesystem path to read from |
|---|---|
| Returns: | SplineTable - the spline surface stored in the given file |
Evaluate a spline surface on a coordinate grid
| Parameters: |
|
|---|---|
| Returns: | an array of values, one for point in the tensor product of the supplied axes |
>>> spline.coefficients.ndim
2
>>> grideval(spline, [[0, 0.5, 1.0], [0.5, 1.0]])
array([[ 1482.110672 , 4529.49084473],
[ 1517.94447213, 4130.34708567],
[ 1506.64602055, 3425.31209966]])
A drop-in replacement for icecube.photospline.glam.glam.grideval().
Note
The bases argument is not supported in this version.
Evaluate the ith nth-order B-spline basis function at x.
| Parameters: |
|
|---|---|
| Returns: | float - the value of the spline at x |
The photospline C library contains a collection of functions for efficiently evaluating the tensor-product B-spline surfaces produced by :py:func`icecube.photospline.spglam.fit` and written to disk as FITS files by icecube.photospline.splinefitstable.write().
A data structure that holds the coefficients, knot grids, and associated metadata needed to evaluate a spline surface:
struct splinetable {
int ndim;
int *order;
double **knots;
long *nknots;
double **extents;
double *periods;
float *coefficients;
long *naxes;
unsigned long *strides;
int naux;
char ***aux;
};
Read a spline table from a FITS file on disk.
| Parameters: |
|
|---|---|
| Returns: | 0 upon success |
Read a spline table from a FITS file in memory.
| Parameters: |
|
|---|
Example:
struct splinetable table;
struct splinetable_buffer buf;
readsplinefitstable("foo.fits", &table);
buf.mem_alloc = &malloc;
buf.mem_realloc = &realloc;
writesplinefitstable_mem(&buf, &table);
splinetable_free(&table);
readsplinefitstable_mem(&buf, &table);
free(buf.data);
/* do stuff with table */
splinetable_free(&table);
Write a spline table to a FITS file on disk.
Write a spline table to a FITS file in memory.
| Parameters: |
|
|---|
Example:
struct splinetable table;
struct splinetable_buffer buf;
readsplinefitstable("foo.fits", &table);
buf.mem_alloc = &malloc;
buf.mem_realloc = &realloc;
writesplinefitstable_mem(&buf, &table);
splinetable_free(&table);
/* do stuff with buf */
free(buf.data);
Get a pointer to the raw string representation of an auxiliary field stored in the FITS header.
| Parameters: |
|
|---|---|
| Returns: | A pointer to the beginning of the value string, or NULL if the key doesn’t exist. |
Parse and store the value of an auxiliary field stored in the FITS header.
| Parameters: |
|
|---|---|
| Returns: | 0 upon success |
Types that can be parsed by splinetable_read_key():
typedef enum {
SPLINETABLE_INT,
SPLINETABLE_DOUBLE
} splinetable_dtype;
Free memory allocated by :c:function:`readsplinefitstable`.
Find the index of the order-0 spline in each dimension that has support at the corresponding coordinate.
| Parameters: |
|
|---|---|
| Returns: | 0 upon success. A non-zero return value indicates that one or more of the coordinates is outside the support of the spline basis in its dimension, in which case the value of the spline surface is 0 by construction. |
Evaluate the spline surface or its derivatives.
| Parameters: |
|
|---|
For example, passing an unset bitmask evaluates the surface:
struct splinetable table;
int centers[table.ndim];
double x[table.ndim];
double v;
if (tablesearchcenters(&table, &x, ¢ers) != 0)
v = ndsplineeval(&table, &x, ¢ers, 0);
else
v = 0;
while setting individual bits evalulates the elements of the gradient:
double gradient[table.ndim];
for (i = 0; i < table.ndim; i++)
gradient[i] = ndsplineeval(&table, &x, ¢ers, (1 << i));
Evaluate the spline surface and all of its derivatives, using SIMD operations to efficiently multiply the same coefficient matrix into multiple bases.
| Parameters: |
|
|---|
On most platforms, each group of 4 bases (e.g. the value and first 3 elements of the gradient) takes the nearly the same number of operations as a single call to ndsplineeval(). As a result, this version is much faster than sequential calls to ndsplineeval() in applications like maximum-likelihood fitting where both the value and entire gradient are required.